Random Selection Of Microorganism ; 8) Staphylococcus aureus ( cocci in clusters )
1)Staphylococcus aureus :
2) Classification :
Domain : Bacteria
Kingdom : Bacteria
Phylum : Firmicutes
Class : Cocci
Order : Bacillales
Family : Staphylococcaceae
Genus : Staphylococcus
Species : Staphylococcus aureus
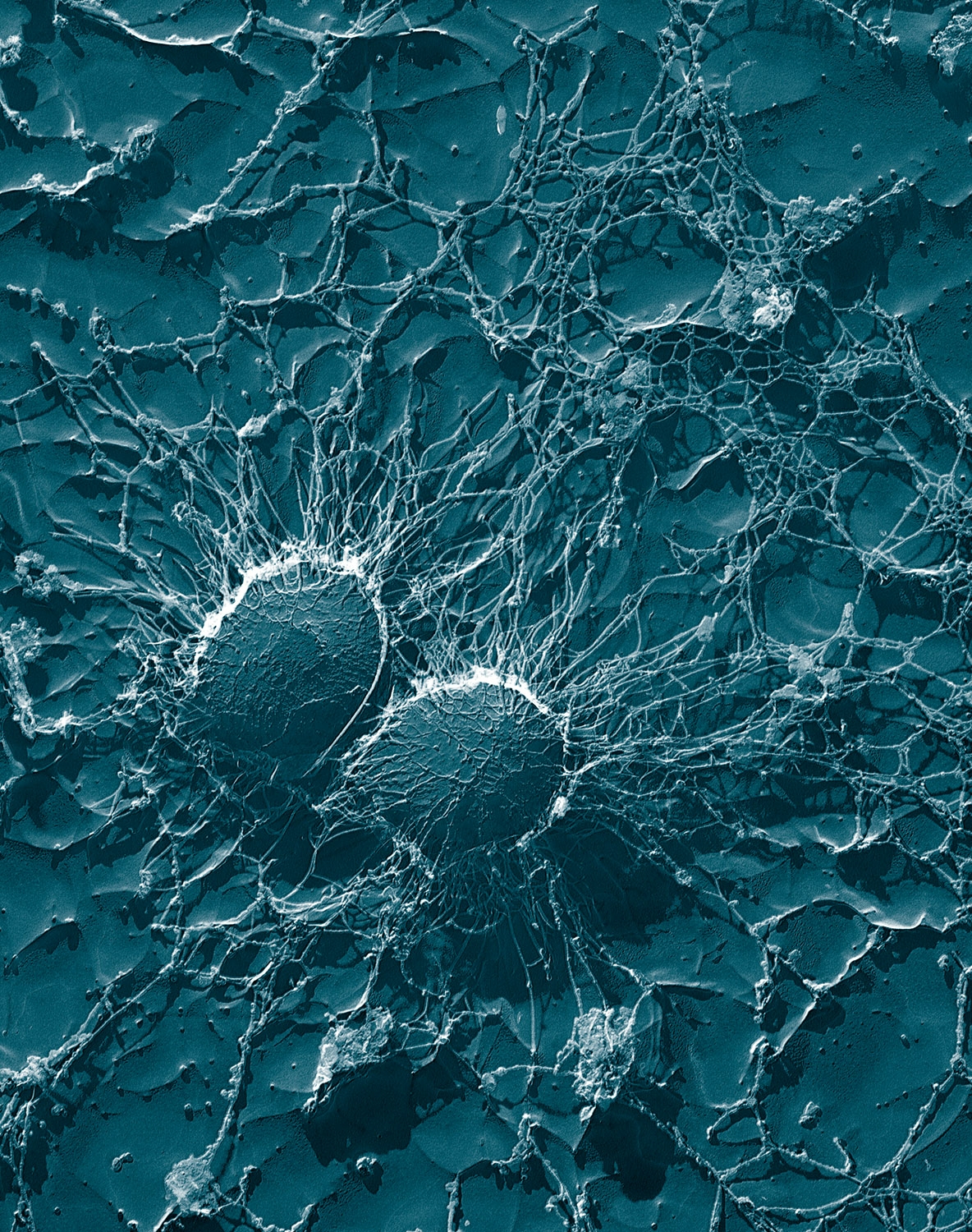
Fig : Observe Cocci in Cluster form ( Bunch )
An estimated 20% to 30% of the human population are long-term carriers S. aureus which can be found as part of the normal skin flora, in the nostrils,and as a normal inhabitant of the lower reproductive tract of women.
S. aureus can cause a range of illnesses, from minor skin infections, such as pimples, impetigo, boils, cellulitis, folliculitis, carbuncles, scalded skin syndrome, and abscesses, to life-threatening diseases such pneumonia, meningitis, osteomyelitis, endocarditis, toxic shock syndrome, bacteremia, and sepsis.
Staphylococcus aureus on basic cultivation media
It is still one of the five most common causes of hospital-acquired infections and is often the cause of wound infections following surgery.
Each year, around 500,000 patients in hospitals of the United States contract a staphylococcal infection, chiefly by S. aureus. Up to 50,000 deaths each year in the USA are linked with S. aureus infections
Introduction :
Staphylococci are spherical gram-positive bacteria, which are immobile and form grape-like clusters. They form bunches because they divide in two planes as opposed to their close relatives streptococci which form chains because they divide only in one plane.
Colonies formed by S. aureus are yellow (thus the name aureus, Latin for gold) and grow large on a rich medium.
Staphylococcus aureus and their genus Staphylococci are facultative anaerobes which means they grow by aerobic respiration or fermentation that produces lactic acid.
As a pathogen, it is important to understand the virulence mechanisms of S. aureus especially the Methicillin-resistant Staphylococcus aureus (MRSA) in order to successfully combat the pathogen.
The increasing population of "super germs" and antibiotic resistant pathogens have increased pressure on researchers to find alternative, more effective ways of fighting these "super germs."
DNA sequencing of this microbe has already isolated the source code of its' resistance to antibiotics, and further research will more than likely lead us to the path of our next artillery against this and many other pathogens.
3) Ecology :
4)Cell Structure & Metabolism:
Staphylococcus aureus is a gram-positive bacteria, which means that the cell wall of this bacteria consists of a very thick peptidoglycan layer. They form spherical colonies in clusters in 2 planes and have no flagella.
Secretions are numerous, but include surface associated adhesins, exoenzymes, and capsular polysaccharides. The capsule is responsible for enhanced virulence of a mucoid strain.
The central routes of glucose metabolism are the Embden-Meyerhof-Parnas (EMP) pathway and the pentose phosphate cycle.
Lactate is the end product of anaerobic glucose metabolism and acetate and CO2 are the products of aerobic growth conditions.
S. aureus can uptake a variety of nutrients including glucose, mannose, mannitol, glucosamine, N-acetylglucosamine, sucrose, lactose, galactose and beta-glucosides.
5)Pathogenicity & Resistance :
Staphylococcus aureus is the most common cause of staph infections and is responsible for various diseases including: mild skin infections (impetigo, folliculitis, etc.), invasive diseases (wound infections, osteomyelitis, bacteremia with metastatic complications, etc.), and toxin mediated diseases (food poisoning, toxic shock syndrome or TSS, scaled skin syndrome, etc.).
Infections are preceded by colonization. Common superficial infections include carbuncles, impetigo, cellulitis, folliculitis.
Community-acquired infections include bacteremia, endocarditis, osteomylitis, pneumonia and wound infections are less common. S. aureus also causes economically important mastitis in cows, sheep and goats.
In the late 1970's an epidemic of toxic shock syndrome (TSS T-1) was brought about by a change in host environment. The environment change was encouraged by a creation of modern technology otherwise known as the super-absorbent tampon.
This new modern convenience product that had swept the nation's females actually created a new nutrient-rich surface area for many bacteria to thrive off of, S. aureus being a major resident.
However, TSS from tampons can be easily avoided by correctly using tampons (read instructions and warning labels listed with tampon products).
MRSA: The first serious emergence of antibiotic resistance staph occurred with a specific strain we refer to as Methicillin-Resistant Staphylococcus aureus, abbreviated as MRSA.
This strain expressed a modified penicillin-binding protein encoded by mecA gene and is present in 4 forms of Staph. Cassette Chromosome. The MRSA, resistant to the antibiotic methicillin, was eventually isolated. Consequently, vancomycin (the most powerful antibiotic in our arsenal) became the primary antibiotic used to combat staphylococcus infection.
In 1997 a strain of S. aureus resistant to vancomycin was isolated, and people are once again exposed to the threat of untreatable staphylococcus infection. MRSA strains are currently a very significant health care problem.
The sequencing of the S. aureus genome will hopefully provide insight into how the organism generates such a variety of toxins, and aid researchers in developing ways of combating the versatile bacterium.
6) Antibiotic Resiatance :
7) Current Research :
Most of the current research on this bacterium involves the proteomics of Staphylococcus aureus and MRSA.
Staph’s resistance to antibiotics has become an increasing problem for today’s society and more research is needed to find our next “super drug”.
There is research focusing on the stress and starvation proteins to predict the physiological state of a cell population
so that we may better understand and find another tactic to combat this bacterium.
Another research based on the proteomics is objectively building a protein expression index for various S. aureus strains and performing comparative analysis of the various strains under different growth conditions.
Methods of achieving this research hold high hopes for development of new techniques.
There is also research being done to investigate hospital acquired MRSA, the link between virulence features of S. aureus, and whether the isolates are from the environment or from patients.
8) Genome :
The Staphylococcus aureus genome, which is the most common species among the Staphylococcus genome projects, is the most completed genome sequence compared to any other microbial species.
The original genome map of Staphylococcus aureus was based on the strain NCTC 8325, initiated by Peter A. Pattee and colleagues. By 2000, the entire genome of strain 8325 had been sequenced and annotated.
Since then, at least six other ‘‘S. aureus’’ strains have been completed (COL, N315, Mu50, MW2, MRSA252, MSSA476).
The Staphylococcus aureus strain NCTC 8325 complete circular genome map shows ~2,900 open reading frames, 61 tRNA genes, 3 structural RNAs, and 5 complete ribosomal RNA operons.
This strain has about 33% G+C content and an average gene length of 824 nucleotides with 85% coding sequence, similar to other S. aureus strains. Half the coding sequence is located predominantly on one replichore and the second half is located predominantly on the other replichore.
Virulence factors are encoded by phages, plasmids, pathogenicity islands and staphylococcus cassette chromosome. Increased resistance for antibiotics is encoded by a transposon (Tn 1546) that was inserted into a conjugated plasmid that also encoded resistance to other things including disinfectants.
MRSA (Methicillin-Resistant Staphylococcus aureus), which is resistant to the antibiotic methicillin, expresses a modified penicillin-binding protein encoded by mecA gene. This was brought about by many evolutions thought horizontal gene transfer of mecA to a wide variety of methicillin susceptible S. aureus strains. The genes for antibiotic resistance in Staphylococcus aureus are located on plasmids or other similar structures.
Diversification within the S. aureus population is achieved through a combination of mutation, recombination and horizontal gene transfer. Evolution of this bacterium can occur through asymptomatic colonization and/or during the course of the caused disease.























Comments
Post a Comment